Abstract
Multiple Myeloma (MM) is an incurable malignancy of plasma cells characterized by wide and remarkable genetic heterogeneity. The amplification of 1q identifies a class of patients characterized by a more aggressive disease course and poor prognosis. While previous research has identified several oncogenes with prognostic relevance located in 1q, the biological mechanisms underlying the poor clinical outcome associated with this alteration is still unclear.
ADAR (Adenosine Deaminase Acting on RNA) is an enzyme responsible for A-to-I editing, a post-transcriptional process which contributes to transcriptome and proteome diversity by conversion of specific adenosines into inosines in both coding and non-coding transcripts. Such changes may affect RNA stability and regulation and introduce mutations in the corresponding proteins.
Recent research has reported dysregulation of RNA editing in cancer, revealing its oncogenicity and clinical relevance. Since ADAR is located in 1q21, the critical amplified region in MM, we sought to investigate the implications of ADAR activity in MM.
We have previously shown increased ADAR expression and A-to-I RNA editing in a small group of newly diagnosed patients with 1q21 amplification enrolled in the MMRF CoMMpass study ( Lagana et al, ASH 2016), and that ADAR expression could differentiate prognosis. To further investigate and characterize the role of ADAR and RNA Editing in MM, we have analyzed 590 RNAseq samples from newly diagnosed patients in CoMMpass. ADAR copy number gain was observed in 111 samples (19%).
ADAR expression was significantly higher in patients with 1q21 gain (p < 0.0001), supporting the role of copy number gain as a driver of ADAR expression. Moreover, patients with high expression of ADAR and no CN alterations, showed significant activation of Interferon pathways (Fig. 1A). This can be explained by the fact that one isoform of ADAR (p150) is regulated by an interferon-inducible promoter. Multivariate regression of ADAR expression against ADAR CN and STAT1 expression, as a measure of interferon signaling activation, showed that both ADAR CN and STAT1 expression were significant independent predictors of ADAR expression.
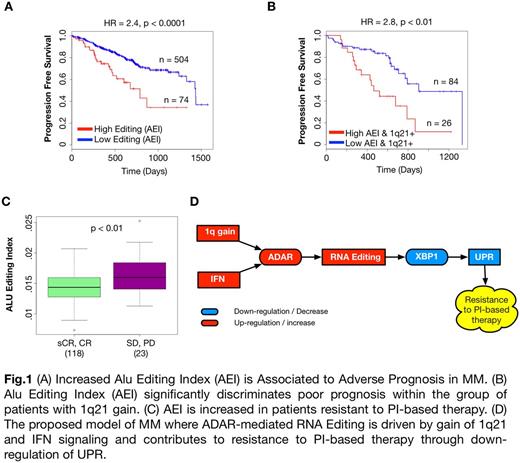
To investigate the functional impact of ADAR activity, we implemented a computational pipeline to calculate sample-wise Alu Editing Index (AEI). A-to-I RNA editing targets predominantly Alu elements, repetitive sequences abundantly interspersed throughout the human genome, mostly within introns and untranslated regions (UTRs). AEI has been developed and validated as a global measure of RNA editing activity at the sample level (Bazak et al, Genome Res 2014). Our analysis in MM revealed that AEI could discriminate between good and poor prognosis in terms of both PFS and OS (p < 0.0001). Moreover, AEI could critically stratify patients with 1q21 gain in terms of PFS (p < 0.01) (Fig. 1B). This finding suggests that RNA editing can be an adverse prognostic factor independently of 1q21 gain.
Patients with high AEI were characterized by down-regulation of immunoglobulin transcripts and of the Unfolded Protein Response (UPR) pathway. In particular, patients with high AEI exhibited down-regulation of key activators of UPR such as NFYC, ATF4 and XBP1, and of transcriptional targets of XBP1s, the active spliced form of XBP1. Decreased UPR and low XBP1s expression in pre-plasmablasts have been shown to induce resistance to proteasome inhibitors (PI) (e.g. Bortezomib) through de-differentiation of plasma cells (Leung-Hagesteijn et al, Cancer Cell 2013). Among patients who received Bortezomib-based first line therapy, we found that AEI was significantly higher (p<0.01) in patients whose best response was stable disease (SD) or progressive disease (PD) (23; 4.5%) compared to patients who had complete (CR) or stringent complete response (sCR) (118; 23%) (Fig. 1C). siRNA meditated knockdown of ADAR increased sensitivity to Bortezomib in MM cell lines with 1q amplification and high ADAR expression (OPM2), consistently with our in-silico findings.
In conclusion, we propose a model of MM where ADAR-mediated RNA editing is driven by gain of 1q21 and interferon signaling and contributes to disease aggressiveness and resistance to PI-based therapy through down-regulation of UPR (Fig. 1D).
Madduri: Foundation Medicine, Inc.: Consultancy. Chari: Millennium Pharmaceuticals, Inc.: Consultancy, Research Funding; Amgen: Honoraria, Research Funding; Novartis: Consultancy, Research Funding; Array BioPharma: Consultancy, Research Funding; Celgene Corporation: Consultancy, Research Funding; Janssen: Consultancy, Research Funding. Cho: Agenus, Inc.: Research Funding; Genentech: Other: advisory board, Research Funding; Ludwig Institute for Cancer Research: Research Funding; Bristol Myers-Squibb: Other: advisory board, Research Funding; Multiple Myeloma Research Foundation: Research Funding. Barlogie: Millenium Pharmaceuticals: Consultancy, Research Funding; Celgene Corporation: Consultancy, Research Funding. Jagannath: Merck: Consultancy; Medicom: Speakers Bureau; Celgene: Consultancy; Bristol-Meyers Squibb: Consultancy; Novartis: Consultancy; MMRF: Speakers Bureau. Dudley: Ecoeos, Inc.: Equity Ownership; Ayasdi, Inc.: Equity Ownership; Ontomics, Inc.: Equity Ownership; Personalis: Patents & Royalties; AstraZeneca: Consultancy; NuMedii, Inc.: Equity Ownership, Patents & Royalties; Janssen Pharmaceuticals, Inc.: Consultancy; GlaxoSmithKline: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal